Sharing Your Building Blocks
Teaching deep learning for bioinformatics.
By Christian TeBordo
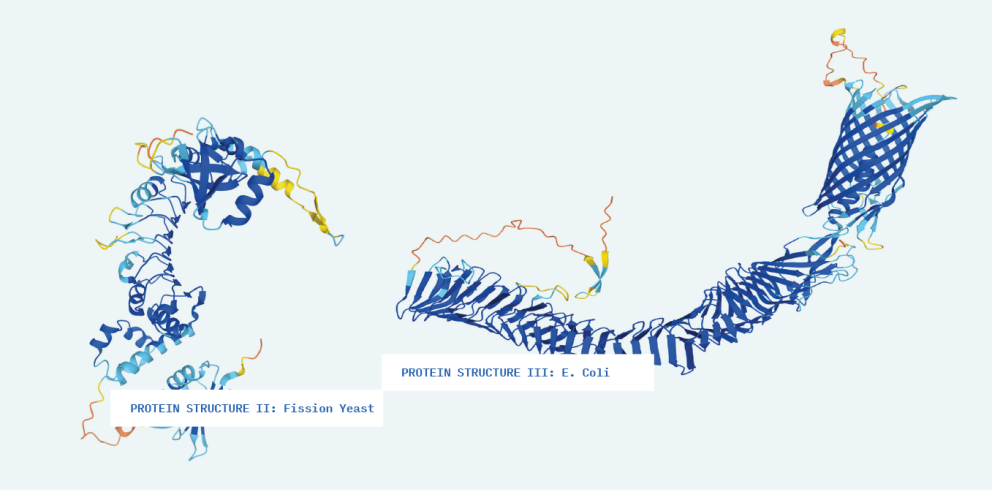
In the spring of 2022, students in Professor Mohammed Zaki’s Machine Learning in Bioinformatics course at Rensselaer Polytechnic Institute were making progress on one of the grand challenges of biology – how to predict which three-dimensional shape a string of amino acids will take as it folds itself into a protein.
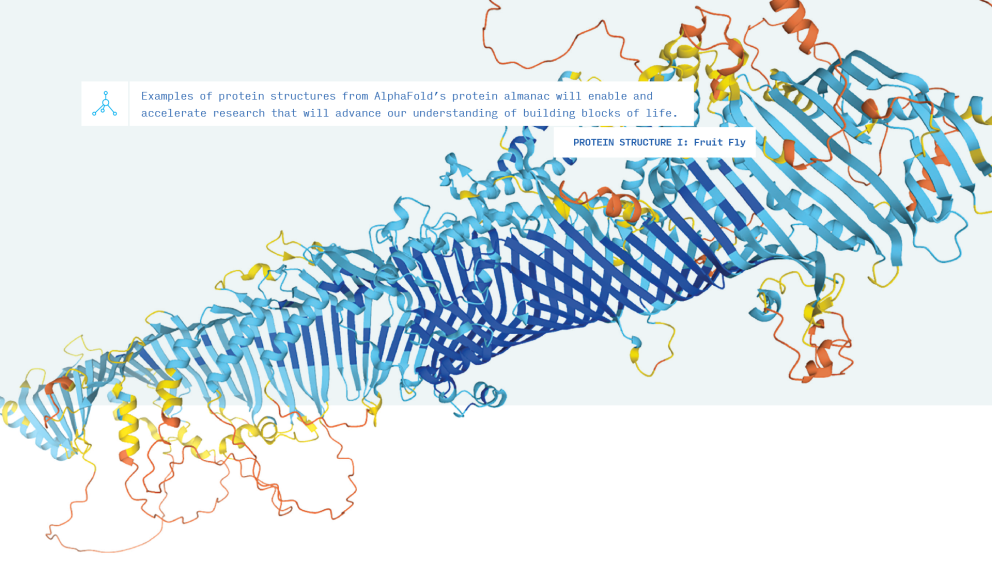
Proteins are often called the building blocks of life, but these building blocks aren’t your standard cubes. There are 21 types of amino acids, which combine into polypeptide chains that can be anywhere from a dozen to thousands long. These chains take an unfathomable variety of shapes through a process sometimes described as “spontaneous origami.”
Science has struggled with the problem of predicting how proteins will fold for more than half a century. Reliable laboratory techniques for exploring protein folding, like X-ray crystallography and nuclear magnetic resonance spectroscopy, are both time-consuming and expensive. For example, there are as many as 400,000 different proteins and protein-variants in the human body, and in 50 years, researchers had managed to identify the structures of fewer than 20 percent of them. Outside the human body, there are another 200 million known proteins, and the count is always increasing.
These numbers sound daunting. Yet the ability to predict what shape a protein will take has implications for everything from medicine to agriculture to climate change. In order to move forward, researchers had to find a new way to determine the structure of these building blocks. Recent advances in machine learning offered a new approach based purely on data.
In November of 2020, AlphaFold, a division of Google’s DeepMind computing group, participated for the second time in the Critical Assessment of Protein Structure Prediction (CASP), a biannual competition in which teams of scientists are assigned amino acid sequences for protein structures that have been confirmed experimentally but are not yet available publicly, and predict those structures with speed and accuracy through computation.
“Our mission at AWS is to make machine learning and data science more accessible to the world.”
Brian Granger, co-creator, Project Jupyter and Jupyter Notebook,
Senior Principal Technologist, AWS
AlphaFold had shown promise in winning the 2018 competition, but its 2020 performance was so successful that the organizers of CASP declared the problem of protein folding effectively solved. Just over a year later, AlphaFold had generated predictions for the entire human proteome.
But there are still shapes to discover and models to refine. To maintain momentum, AlphaFold made the source code available for free, along with details about the research methodology. This is where Professor Zaki’s class comes in. His students would explore the solution to a problem that seemed almost insurmountable only a few years ago. They studied all the deep learning building blocks required to implement AlphaFold on their own.
With its extensive resources in biotechnology and machine learning, investment in supercomputing power, and culture of cross-disciplinary collaboration, Rensselaer is uniquely suited to participating in these revolutionary developments. AiMOS, the Artificial Intelligence Multiprocessing Optimized System, at the Rensselaer Center for Computational Innovations, is the most powerful supercomputer housed on a university campus. This product of a collaboration between the university, IBM, Empire State Development, and NY CREATES is capable of doing one trillion calculations per second.
“The supercomputer is important for large-scale simulations, and we had access to it,” says Zaki, professor of computer science. “But AiMOS has a steep learning curve. So while the students were getting used to that, we wanted another way to get started.”
Zaki approached Amazon Web Services (AWS) to see if they could help. Amazon SageMaker is considered a leader in machine learning platforms, enabling customers to manage the entire lifecycle from data preparation, model building, training and tuning, and deployment and management. AWS had recently released SageMaker Studio Lab, which offers a simple user interface for developers to experiment with building and training models. It provided each of Zaki’s students with a powerful GPU environment, as well as dedicated storage that allowed them to save their work between sessions. Best of all, it was completely free.
“Our mission at AWS is to make machine learning and data science more accessible to the world,” says Brian Granger, co-creator of Project Jupyter and the Jupyter Notebook and Senior Principal Technologist at AWS. “Amazon SageMaker Studio Lab provides everything you need to explore this space: compute, persistent storage, a Jupyter notebook-based IDE, and the ability to install the open-source packages you need to get work done. All this is free and only requires an email address to create an account. Professor Zaki’s course shows how much you can get done with these simple building blocks – it is delightful to see.”
This collaboration also helped Amazon fine-tune the service as it prepared for the official launch. The students’ sustained work in the Studio Lab allowed them to observe the way consistent and dedicated users might utilize it, which inspired new features like event management “referral codes” that prioritize users who are part of trusted events, such as machine learning classes.
“In the final capstone we separated into groups of two students each, and they implemented the AlphaFold protein folding approach and tested it out. The idea was to teach them all the techniques for protein folding and some other machine learning problems in bioinformatics.”
Mohammed J. Zaki, PH.D., Department Head, Computer Science
Zaki’s course was a success. “In the final capstone we separated into groups of two students each, and they implemented the AlphaFold protein folding approach and tested it out,” he says. “The idea was to teach them all the techniques for protein folding and some other machine learning problems in bioinformatics. They were really happy to see and understand what’s going on under the hood.”
In the end, it was a collaboration between one class at Rensselaer, the state of New York, several of the world’s largest corporations, and interdisciplinary researchers from all over the globe, a testament to what’s possible when scientists build together.